# load libraries
library(GO.db);
library(AnnotationDbi);
library(clusterProfiler);
library(org.Dm.eg.db,verbose=F,quietly=T);
library(ggplot2)
fly gene ID mapping via the ‘select’ function
# which kinds of data are retrievable via `select`
columns(org.Dm.eg.db)
## [1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
## [6] "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "FLYBASE"
## [11] "FLYBASECG" "FLYBASEPROT" "GENENAME" "GENETYPE" "GO"
## [16] "GOALL" "MAP" "ONTOLOGY" "ONTOLOGYALL" "PATH"
## [21] "PMID" "REFSEQ" "SYMBOL" "UNIPROT"
# use keys as query to extract other column information
set.seed(123)
(k=sample(keys(org.Dm.eg.db,keytype='SYMBOL'),5))
## [1] "CG7215" "CG33964" "Pkd2" "Eaat2" "amos"
AnnotationDbi::select(org.Dm.eg.db,keys=k,keytype="SYMBOL",c("FLYBASE","GENENAME"))
## SYMBOL FLYBASE GENENAME
## 1 CG7215 FBgn0038571 uncharacterized protein
## 2 CG33964 FBgn0053964 uncharacterized protein
## 3 Pkd2 FBgn0041195 Polycystic kidney disease 2
## 4 Eaat2 FBgn0026438 Excitatory amino acid transporter 2
## 5 amos FBgn0003270 absent MD neurons and olfactory sensilla
extracting all fly genes and their annotated GO terms
all.fly.genes<-keys(org.Dm.eg.db,"FLYBASE")
length(all.fly.genes)
## [1] 25097
fb2go=AnnotationDbi::select(org.Dm.eg.db,keys=all.fly.genes,keytype = 'FLYBASE',
columns = c('GO'))
head(fb2go)
## FLYBASE GO EVIDENCE ONTOLOGY
## 1 FBgn0040373 GO:0000139 IBA CC
## 2 FBgn0040373 GO:0006486 IEA BP
## 3 FBgn0040373 GO:0016757 IBA MF
## 4 FBgn0040373 GO:0016758 IEA MF
## 5 FBgn0040372 GO:0002039 IEA MF
## 6 FBgn0040372 GO:0002165 IMP BP
x<-split(fb2go$GO,f=fb2go$FLYBASE)
x[c(1,2,3)]
## $FBgn0000001
## [1] NA
##
## $FBgn0000003
## [1] "GO:0005786" "GO:0006614"
##
## $FBgn0000008
## [1] "GO:0003674" "GO:0005912" "GO:0016324" "GO:0048749"
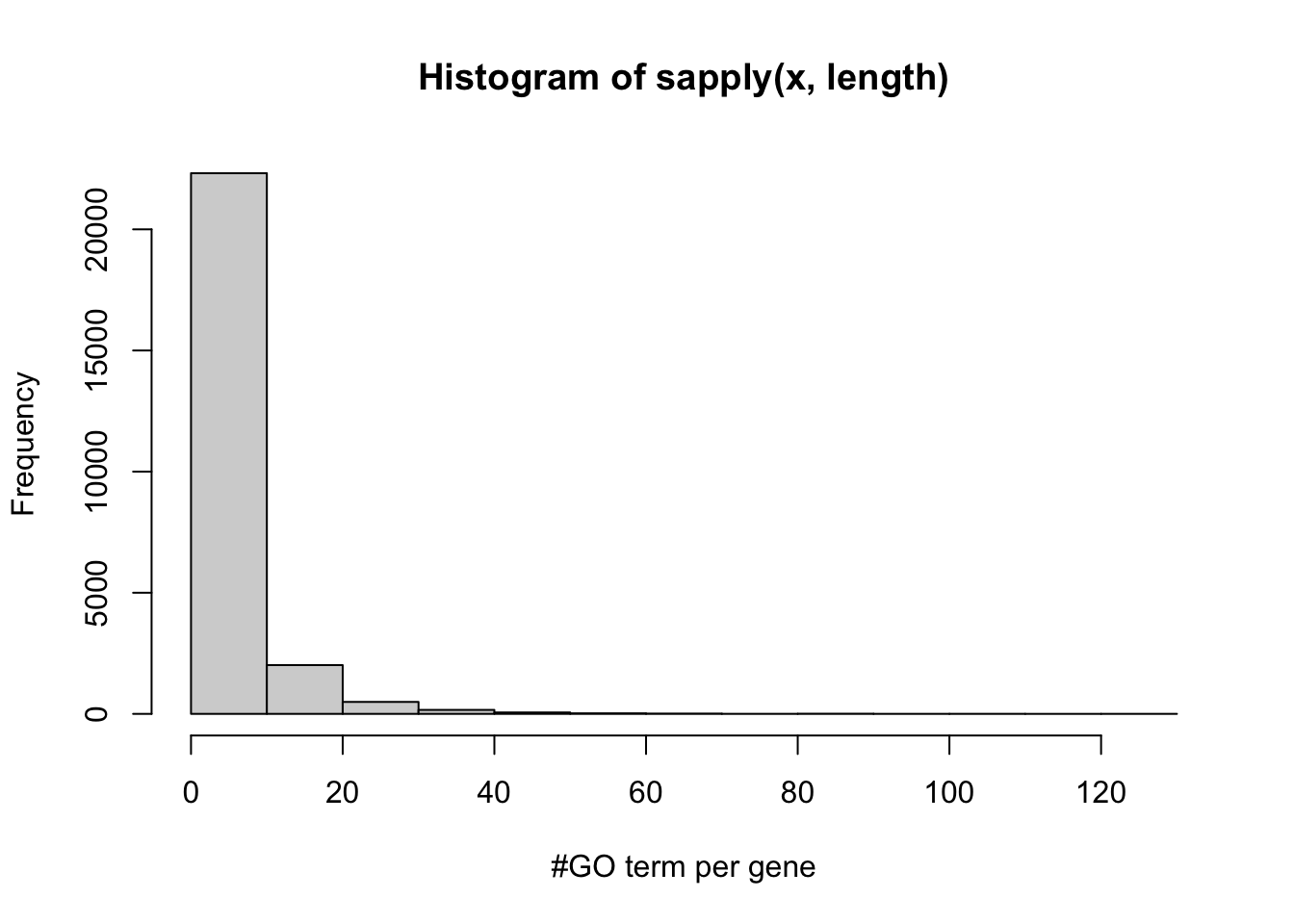
hist(sapply(x,length),xlab='#GO term per gene')
go2fb <- split( fb2go$FLYBASE,f=as.factor( fb2go$GO) )
str(go2fb[1:3], vec.len=3)
## List of 3
## $ GO:0000001: chr [1:3] "FBgn0029891" "FBgn0033690" "FBgn0261618"
## $ GO:0000002: chr [1:6] "FBgn0032154" "FBgn0040268" "FBgn0010438" ...
## $ GO:0000003: chr [1:4] "FBgn0023509" "FBgn0003742" "FBgn0003742" ...
retrieve GO term full descriptions
# GO.db usaage info: https://www.bioconductor.org/packages/release/bioc/vignettes/annotate/inst/doc/GOusage.pdf
# which can be used by `select`
columns(GO.db)
## [1] "DEFINITION" "GOID" "ONTOLOGY" "TERM"
set.seed(111)
go.id<-sample(keys(GO.db,keytype = 'GOID'),5);
AnnotationDbi::select(GO.db,keys=go.id,keytype ='GOID',
columns=c("DEFINITION","ONTOLOGY","TERM"));
## GOID
## 1 GO:0099549
## 2 GO:0060974
## 3 GO:0104004
## 4 GO:0009155
## 5 GO:1902586
## DEFINITION
## 1 Cell-cell signaling between presynapse and postsynapse mediated by carbon monoxide.
## 2 The orderly movement of a cell from one site to another that contribute to the formation of the heart. The initial heart structure is made up of mesoderm-derived heart progenitor cells and neural crest-derived cells.
## 3 Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an environmental stimulus.
## 4 The chemical reactions and pathways resulting in the breakdown of purine deoxyribonucleotide, a compound consisting of deoxyribonucleoside (a purine base linked to a deoxyribose sugar) esterified with a phosphate group at either the 3' or 5'-hydroxyl group of the sugar.
## 5 <NA>
## ONTOLOGY TERM
## 1 BP trans-synaptic signaling by carbon monoxide
## 2 BP cell migration involved in heart formation
## 3 BP cellular response to environmental stimulus
## 4 BP purine deoxyribonucleotide catabolic process
## 5 BP multi-organism intercellular transport
GO enrichment analysis and visualization
test.genes=c("ATPsynC","ATPsyngamma","blw","COX6B","COX7A","Cyt-c-p","porin","sesB");
gene.df <- clusterProfiler::bitr(test.genes, fromType = "SYMBOL",
toType = c("ENTREZID","FLYBASE","GENENAME"),
OrgDb = org.Dm.eg.db)
ego <- enrichGO(gene = gene.df$ENTREZID,
OrgDb = org.Dm.eg.db,
#keyType = 'SYMBOL',
ont = "BP",
#ont = "MF",
#ont = "CC",
pAdjustMethod = "BH",
pvalueCutoff = 0.05,
qvalueCutoff = 0.05,
readable = TRUE)
#use simplify to remove redudant GO terms, a larger cutoff leads to a smaller number of returned GO terms
#ref: https://guangchuangyu.github.io/2015/10/use-simplify-to-remove-redundancy-of-enriched-go-terms/
x<-simplify(ego, cutoff=0.7, by="p.adjust", select_fun=min)
result<-x@result[order(x@result$p.adjust),]
x1=result;
x1$GeneRatio1=x1$GeneRatio
x1$GeneRatio=sapply(x1$GeneRatio,function(x){
p=as.numeric(unlist(strsplit(x,'/')))
p[1]/p[2]
})
summary(x1$GeneRatio)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.1250 0.1250 0.2500 0.2939 0.5000 0.7500
x1=x1[order(x1$p.adjust),]
x1$desp=factor(x1$Description,levels=rev(x1$Description))
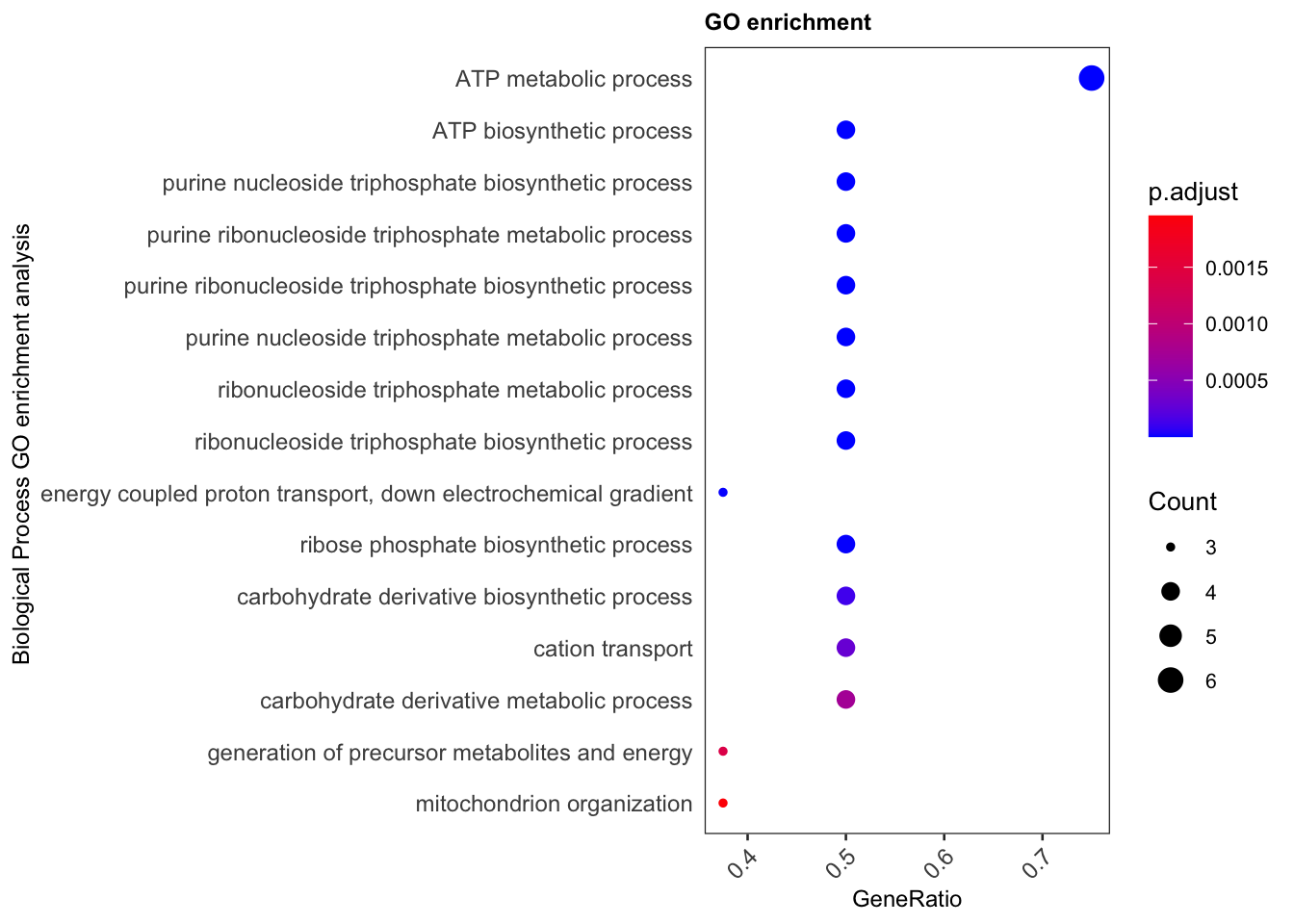
ggplot(subset(x1,Count>=3),aes(x=GeneRatio,y=desp,size=Count,col=p.adjust))+
geom_point()+theme_bw(base_size=10)+
scale_color_gradient(low="blue", high="red")+
scale_size(range = c(1,4))+
ylab('Biological Process GO enrichment analysis ')+
ggtitle('GO enrichment')+
theme(
plot.title =element_text(size=9, face='bold'),
panel.grid = element_blank(),
axis.text=element_text(size=9),
axis.title=element_text(size=9),
axis.text.x=element_text(size=9,angle=45, hjust=1),
axis.text.y=element_text(size=9,angle=0, hjust=1),
axis.ticks.y = element_blank())
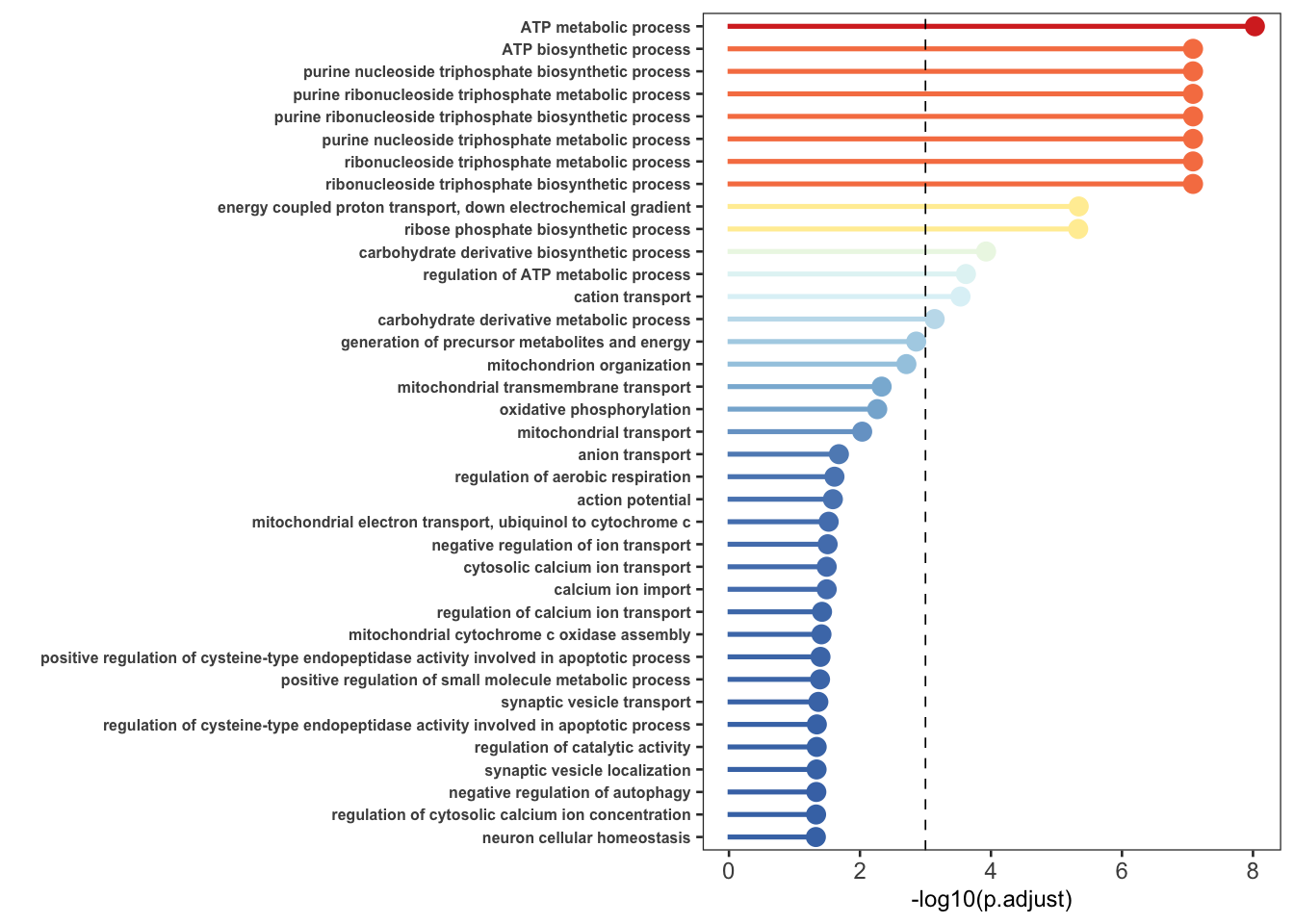
p.adj.cutoff=0.001;
p.line = -1*log(p.adj.cutoff,base=10)
x1$log.p.adjust = -1*log(x1$p.adjust,base=10)
ggplot(x1,aes(x=desp,y=log.p.adjust,col=log.p.adjust))+
geom_bar(aes(fill=log.p.adjust),stat='identity',width = 0.1)+
theme_bw(base_size=10)+ylab('-log10(p.adjust)')+xlab('')+
scale_color_distiller(name='',palette = "RdYlBu")+
scale_fill_distiller(name='',palette = "RdYlBu")+
#scale_color_distiller(name='',palette = "Dark2")+
geom_point(size=3)+coord_flip()+
geom_hline(yintercept = p.line, linetype="dashed",
color = "black", size=0.3)+
theme(legend.position = 'none',
axis.title = element_text(size=9),
axis.text.y=element_text(size=6,face="bold"),
axis.text.x=element_text(size=9),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
devtools::session_info()
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Big Sur/Monterey 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/Los_Angeles
## date 2023-05-26
## pandoc 2.18 @ /Applications/RStudio.app/Contents/MacOS/quarto/bin/tools/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## AnnotationDbi * 1.56.2 2021-11-09 [1] Bioconductor
## ape 5.6-2 2022-03-02 [1] CRAN (R 4.1.2)
## aplot 0.1.2 2022-01-10 [1] CRAN (R 4.1.2)
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.1.0)
## Biobase * 2.54.0 2021-10-26 [1] Bioconductor
## BiocGenerics * 0.40.0 2021-10-26 [1] Bioconductor
## BiocParallel 1.28.3 2021-12-09 [1] Bioconductor
## Biostrings 2.62.0 2021-10-26 [1] Bioconductor
## bit 4.0.4 2020-08-04 [1] CRAN (R 4.1.0)
## bit64 4.0.5 2020-08-30 [1] CRAN (R 4.1.0)
## bitops 1.0-7 2021-04-24 [1] CRAN (R 4.1.0)
## blob 1.2.2 2021-07-23 [1] CRAN (R 4.1.0)
## blogdown 1.17.2 2023-05-27 [1] Github (rstudio/blogdown@75e4855)
## bookdown 0.25 2022-03-16 [1] CRAN (R 4.1.2)
## brio 1.1.3 2021-11-30 [1] CRAN (R 4.1.0)
## bslib 0.3.1 2021-10-06 [1] CRAN (R 4.1.0)
## cachem 1.0.6 2021-08-19 [1] CRAN (R 4.1.0)
## callr 3.7.0 2021-04-20 [1] CRAN (R 4.1.0)
## cli 3.5.0 2022-12-20 [1] CRAN (R 4.1.2)
## clusterProfiler * 4.2.2 2022-01-13 [1] Bioconductor
## colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.1.0)
## crayon 1.5.0 2022-02-14 [1] CRAN (R 4.1.2)
## data.table 1.14.2 2021-09-27 [1] CRAN (R 4.1.0)
## DBI 1.1.2 2021-12-20 [1] CRAN (R 4.1.0)
## desc 1.4.0 2021-09-28 [1] CRAN (R 4.1.0)
## devtools 2.4.3 2021-11-30 [1] CRAN (R 4.1.0)
## digest 0.6.29 2021-12-01 [1] CRAN (R 4.1.0)
## DO.db 2.9 2022-03-12 [1] Bioconductor
## DOSE 3.20.1 2021-11-18 [1] Bioconductor
## downloader 0.4 2015-07-09 [1] CRAN (R 4.1.0)
## dplyr 1.0.8 2022-02-08 [1] CRAN (R 4.1.2)
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.0)
## enrichplot 1.14.2 2022-02-24 [1] Bioconductor
## evaluate 0.15 2022-02-18 [1] CRAN (R 4.1.2)
## fansi 1.0.2 2022-01-14 [1] CRAN (R 4.1.2)
## farver 2.1.0 2021-02-28 [1] CRAN (R 4.1.0)
## fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
## fastmatch 1.1-3 2021-07-23 [1] CRAN (R 4.1.0)
## fgsea 1.20.0 2021-10-26 [1] Bioconductor
## fs 1.5.2 2021-12-08 [1] CRAN (R 4.1.0)
## generics 0.1.2 2022-01-31 [1] CRAN (R 4.1.2)
## GenomeInfoDb 1.30.1 2022-01-30 [1] Bioconductor
## GenomeInfoDbData 1.2.7 2022-02-19 [1] Bioconductor
## ggforce 0.3.3 2021-03-05 [1] CRAN (R 4.1.0)
## ggfun 0.0.5 2022-01-20 [1] CRAN (R 4.1.2)
## ggplot2 * 3.4.0 2022-11-04 [1] CRAN (R 4.1.2)
## ggplotify 0.1.0 2021-09-02 [1] CRAN (R 4.1.0)
## ggraph 2.0.5 2021-02-23 [1] CRAN (R 4.1.0)
## ggrepel 0.9.1 2021-01-15 [1] CRAN (R 4.1.0)
## ggtree 3.2.1 2021-11-16 [1] Bioconductor
## glue 1.6.1 2022-01-22 [1] CRAN (R 4.1.2)
## GO.db * 3.14.0 2022-03-12 [1] Bioconductor
## GOSemSim 2.20.0 2021-10-26 [1] Bioconductor
## graphlayouts 0.8.0 2022-01-03 [1] CRAN (R 4.1.2)
## gridExtra 2.3 2017-09-09 [1] CRAN (R 4.1.0)
## gridGraphics 0.5-1 2020-12-13 [1] CRAN (R 4.1.0)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 4.1.0)
## highr 0.9 2021-04-16 [1] CRAN (R 4.1.0)
## htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
## httr 1.4.2 2020-07-20 [1] CRAN (R 4.1.0)
## igraph 1.2.11 2022-01-04 [1] CRAN (R 4.1.2)
## IRanges * 2.28.0 2021-10-26 [1] Bioconductor
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.1.0)
## jsonlite 1.8.0 2022-02-22 [1] CRAN (R 4.1.2)
## KEGGREST 1.34.0 2021-10-26 [1] Bioconductor
## knitr 1.37 2021-12-16 [1] CRAN (R 4.1.0)
## labeling 0.4.2 2020-10-20 [1] CRAN (R 4.1.0)
## lattice 0.20-45 2021-09-22 [1] CRAN (R 4.1.3)
## lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.1.0)
## lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.1.2)
## magrittr 2.0.2 2022-01-26 [1] CRAN (R 4.1.2)
## MASS 7.3-55 2022-01-16 [1] CRAN (R 4.1.3)
## Matrix 1.4-0 2021-12-08 [1] CRAN (R 4.1.3)
## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.1.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.1.0)
## nlme 3.1-155 2022-01-16 [1] CRAN (R 4.1.3)
## org.Dm.eg.db * 3.14.0 2022-03-12 [1] Bioconductor
## patchwork 1.1.1 2020-12-17 [1] CRAN (R 4.1.0)
## pillar 1.7.0 2022-02-01 [1] CRAN (R 4.1.2)
## pkgbuild 1.3.1 2021-12-20 [1] CRAN (R 4.1.0)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.0)
## pkgload 1.2.4 2021-11-30 [1] CRAN (R 4.1.0)
## plyr 1.8.6 2020-03-03 [1] CRAN (R 4.1.0)
## png 0.1-7 2013-12-03 [1] CRAN (R 4.1.0)
## polyclip 1.10-0 2019-03-14 [1] CRAN (R 4.1.0)
## prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.1.0)
## processx 3.5.2 2021-04-30 [1] CRAN (R 4.1.0)
## ps 1.6.0 2021-02-28 [1] CRAN (R 4.1.0)
## purrr 0.3.4 2020-04-17 [1] CRAN (R 4.1.0)
## qvalue 2.26.0 2021-10-26 [1] Bioconductor
## R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
## RColorBrewer 1.1-2 2014-12-07 [1] CRAN (R 4.1.0)
## Rcpp 1.0.8 2022-01-13 [1] CRAN (R 4.1.2)
## RCurl 1.98-1.6 2022-02-08 [1] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [1] CRAN (R 4.1.0)
## reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.1.0)
## rlang 1.0.6 2022-09-24 [1] CRAN (R 4.1.2)
## rmarkdown 2.13 2022-03-10 [1] CRAN (R 4.1.2)
## rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.1.0)
## RSQLite 2.2.10 2022-02-17 [1] CRAN (R 4.1.2)
## rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.0)
## S4Vectors * 0.32.3 2021-11-21 [1] Bioconductor
## sass 0.4.0 2021-05-12 [1] CRAN (R 4.1.0)
## scales 1.2.1 2022-08-20 [1] CRAN (R 4.1.2)
## scatterpie 0.1.7 2021-08-20 [1] CRAN (R 4.1.0)
## sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.1.0)
## shadowtext 0.1.1 2022-01-10 [1] CRAN (R 4.1.2)
## stringi 1.7.8 2022-07-11 [1] CRAN (R 4.1.2)
## stringr 1.4.0 2019-02-10 [1] CRAN (R 4.1.0)
## testthat 3.1.2 2022-01-20 [1] CRAN (R 4.1.2)
## tibble 3.1.6 2021-11-07 [1] CRAN (R 4.1.0)
## tidygraph 1.2.0 2020-05-12 [1] CRAN (R 4.1.0)
## tidyr 1.2.0 2022-02-01 [1] CRAN (R 4.1.2)
## tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.1.0)
## tidytree 0.3.9 2022-03-04 [1] CRAN (R 4.1.2)
## treeio 1.18.1 2021-11-14 [1] Bioconductor
## tweenr 1.0.2 2021-03-23 [1] CRAN (R 4.1.0)
## usethis 2.1.5 2021-12-09 [1] CRAN (R 4.1.0)
## utf8 1.2.2 2021-07-24 [1] CRAN (R 4.1.0)
## vctrs 0.5.1 2022-11-16 [1] CRAN (R 4.1.2)
## viridis 0.6.2 2021-10-13 [1] CRAN (R 4.1.0)
## viridisLite 0.4.0 2021-04-13 [1] CRAN (R 4.1.0)
## withr 2.5.0 2022-03-03 [1] CRAN (R 4.1.2)
## xfun 0.39 2023-04-20 [1] CRAN (R 4.1.2)
## XVector 0.34.0 2021-10-26 [1] Bioconductor
## yaml 2.3.4 2022-02-17 [1] CRAN (R 4.1.2)
## yulab.utils 0.0.4 2021-10-09 [1] CRAN (R 4.1.0)
## zlibbioc 1.40.0 2021-10-26 [1] Bioconductor
##
## [1] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────


